Time-structure Independent Components Analysis (tICA)
Background
Time-structure independent components analysis (tICA) is a method for finding the slowest-relaxing degrees of freedom
in a time series data set which can be formed from linear combinations from a set of input degrees of freedom.
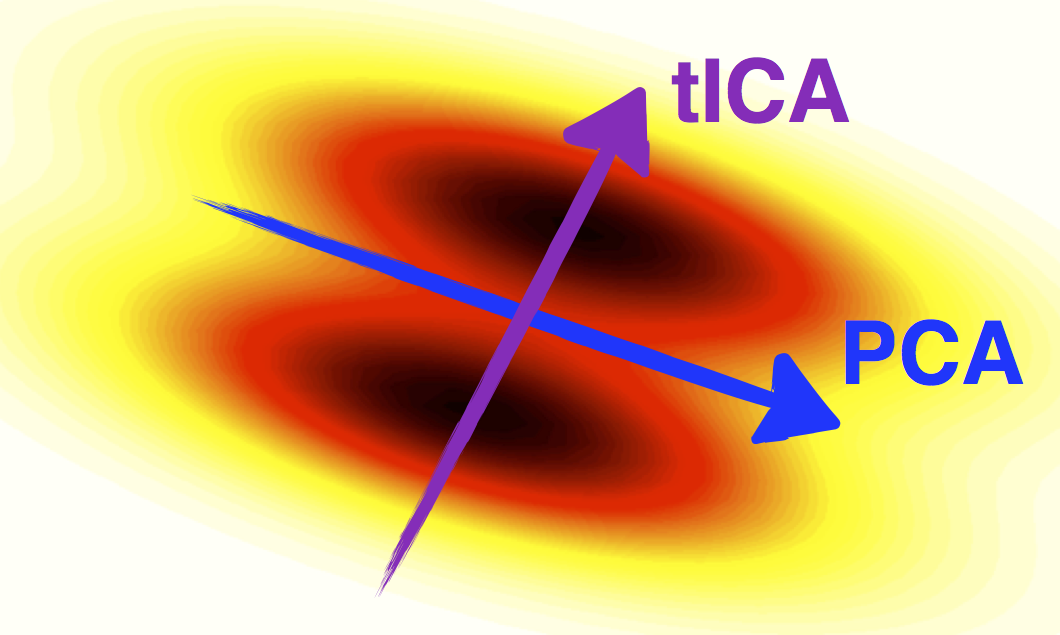
tICA can be used as a dimensionality reduction method, and in that capacity it is somewhat similar to PCA.
However whereas PCA finds high-variance linear combinations of the input degrees of freedom, tICA finds
high-autocorrelation linear combinations of the input degrees of freedom.
Algorithms
| tICA([n_components, lag_time, gamma, ...]) |
Time-structure Independent Component Analysis (tICA) |
| SparseTICA(n_components[, lag_time, gamma, ...]) |
Sparse time-structure Independent Component Analysis (tICA). |
Combination with MSM
While the tICs are themselves approximations to the dominant eigenfunctions
of the propagator / transfer operator, the approach taken in and
is to “stack” tICA with Markov state models (MSMs). For example, in , Perez-Hernandez et
al. first measured the 66 atom-atom distances between a set of atoms in each
frame of their MD trajectories, and then use tICA to find the slowest 1, 4, and
10 linear combinations of these degrees of freedom and transform the
66-dimensional dataset into a 1, 4, or 10-dimensional dataset. Then, they apply
KMeans to the resulting data and build an MSM.
Example
from msmbuilder.decomposition import tICA
from msmbuilder.cluster import KMeans
from msmbuilder.msm import MarkovStateModel
from sklearn.pipeline import Pipeline
pipeline = Pipeline([
('tica', tICA(n_components=4)),
('kmeans', KMeans(n_clusters=1000)),
('msm': MarkovStateModel(),
])
# load a list of 2D arrays, each of shape (length_of_trajectory, n_features)
dataset = ...
pipeline.fit(dataset)
References