Mutual Information¶
Imports¶
In [1]:
%matplotlib inline
import numpy as np
from sklearn.pipeline import Pipeline
from msmbuilder.example_datasets import FsPeptide
from msmbuilder.featurizer import DihedralFeaturizer
from msmbuilder.preprocessing import RobustScaler
from msmbuilder.decomposition import tICA
from msmbuilder.cluster import KMeans
from msmbuilder.msm import MarkovStateModel
from msmbuilder.tpt import net_fluxes, paths
import mdtraj as md
from matplotlib.colors import rgb2hex
from nglview import MDTrajTrajectory, NGLWidget
import msmexplorer as msme
from mdentropy.metrics import DihedralMutualInformation
rs = np.random.RandomState(42)
Load Trajectories¶
In [2]:
trajectories = FsPeptide(verbose=False).get().trajectories
Build Markov Model¶
In [3]:
pipeline = Pipeline([
('dihedrals', DihedralFeaturizer()),
('scaler', RobustScaler()),
('tica', tICA(n_components=2, lag_time=10)),
('kmeans', KMeans(n_clusters=12, random_state=rs)),
('msm', MarkovStateModel(lag_time=1))
])
msm_assignments = pipeline.fit_transform(trajectories)
msm = pipeline.get_params()['msm']
Identify Top Folding Pathway¶
In [4]:
sources, sinks = [msm.populations_.argmin()], [msm.populations_.argmax()]
net_flux = net_fluxes(sources, sinks, msm)
paths, _ = paths(sources, sinks, net_flux, num_paths=0)
samples = msm.draw_samples(msm_assignments, n_samples=1000, random_state=rs)
xyz = []
for state in paths[0]:
for traj_id, frame in samples[state]:
xyz.append(trajectories[traj_id][frame].xyz)
pathway = md.Trajectory(np.concatenate(xyz, axis=0), trajectories[0].topology)
pathway.superpose(pathway[0])
Out[4]:
Calculate Mutual information¶
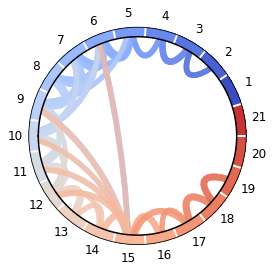
In [5]:
dmutinf = DihedralMutualInformation(n_bins=3, method='knn', normed=True)
M = dmutinf.partial_transform(pathway)
M -= M.diagonal() * np.eye(*M.shape)
labels = [str(res.index) for res in trajectories[0].topology.residues
if res.name not in ['ACE', 'NME']]
ax = msme.plot_chord(M, threshold=.5, labels=labels,)
In [6]:
from nglview import MDTrajTrajectory, NGLWidget
t = MDTrajTrajectory(pathway)
view = NGLWidget(t)
view
In [7]:
scores = np.real(np.linalg.eig(M)[1][0])
scores -= scores.min()
scores /= scores.max()
cmap = msme.utils.make_colormap(['rawdenim', 'lightgrey', 'pomegranate'])
reslist = [str(res.index) for res in pathway.topology.residues][1:-1]
view.clear()
view.clear_representations()
view.add_cartoon('protein', color='white')
for i, color in enumerate(cmap(scores)):
view.add_representation('ball+stick', reslist[i], color=rgb2hex(color))
view.camera = 'orthographic'
(mutual-information.ipynb; mutual-information.eval.ipynb; mutual-information.py)