Ward Clustering¶
Ward Clustering¶
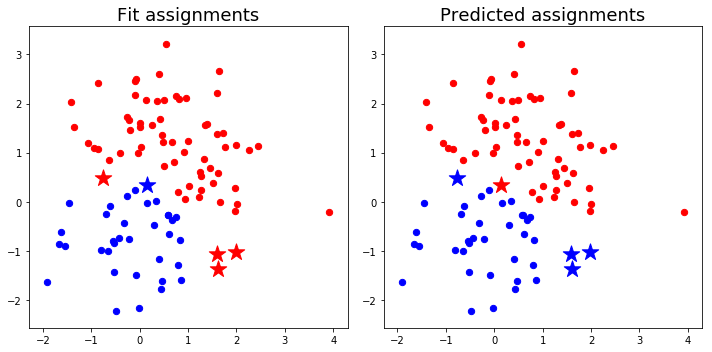
We fit some random points to 2 clusters using the Ward metric and then predict their cluster assignments using the new prediction function. Due to the cardinality dependence of the Ward objective function, data points at the edges of clusters may be assigned to different clusters than the ones to which they were fit.
Generate some random data¶
In [1]:
%matplotlib inline
from matplotlib import pyplot as plt
import numpy as np
xy1 = np.random.randn(50,2)
xy2 = np.random.randn(50,2)+1
xy = np.concatenate([xy1,xy2])
plt.scatter(xy[:,0], xy[:,1])
plt.tight_layout()
Cluster with Ward clustering¶
In [2]:
from msmbuilder.cluster import LandmarkAgglomerative
clusterer = LandmarkAgglomerative(
n_clusters=2, n_landmarks=None,
linkage='ward', metric='euclidean')
clusterer.fit([xy])
fit_assignments = clusterer.landmark_labels_
predict_assignments = clusterer.predict([xy])[0]
Investigate fit/predict fidelity¶
In [3]:
count = np.sum(fit_assignments == predict_assignments)
print("Prediction maintains {}% fidelity to fit assignments."
.format(100*count/(xy.shape[0])))
In [4]:
discrep_list = np.where(fit_assignments != predict_assignments)[0]
discrep_list
Out[4]:
Group fit and predict points by cluster assignments¶
In [5]:
fit_0 = xy[fit_assignments == 0]
fit_1 = xy[fit_assignments == 1]
pred_0 = xy[predict_assignments == 0]
pred_1 = xy[predict_assignments == 1]
c_fit_list = fit_assignments[discrep_list]
c_pred_list = predict_assignments[discrep_list]
Visualize clustering results¶
Highlight discrepancies between fit and predict
In [6]:
fig = plt.figure(figsize=(10,5))
ax1 = plt.subplot(1,2,1)
plt.title('Fit assignments',fontsize=18)
plt.scatter(fit_0[:,0],fit_0[:,1],c='b',s=40)
plt.scatter(fit_1[:,0],fit_1[:,1],c='r',s=40)
xy_star = xy[discrep_list[c_fit_list==0]]
plt.scatter(xy_star[:,0], xy_star[:,1], c='b', s=300, marker='*')
xy_star = xy[discrep_list[c_fit_list==1]]
plt.scatter(xy_star[:,0], xy_star[:,1], c='r', s=300, marker='*')
plt.subplot(1,2,2, sharex=ax1, sharey=ax1)
plt.title('Predicted assignments',fontsize=18)
plt.scatter(pred_0[:,0],pred_0[:,1],c='b',s=40)
plt.scatter(pred_1[:,0],pred_1[:,1],c='r',s=40)
xy_star = xy[discrep_list[c_pred_list==0]]
plt.scatter(xy_star[:,0], xy_star[:,1], c='b', s=300, marker='*')
xy_star = xy[discrep_list[c_pred_list==1]]
plt.scatter(xy_star[:,0], xy_star[:,1], c='r', s=300, marker='*')
plt.tight_layout()
(Ward-Clustering.ipynb; Ward-Clustering.eval.ipynb; Ward-Clustering.py)