tICA vs. PCA¶
tICA vs. PCA¶
This example uses OpenMM to generate example data to compare two methods for dimensionality reduction: tICA and PCA.
Define dynamics¶
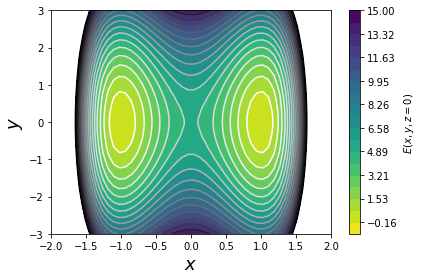
First, let's use OpenMM to run some dynamics on the 3D potential energy function
$$E(x,y,z) = 5 \cdot (x-1)^2 \cdot (x+1)^2 + y^2 + z^2$$
From looking at this equation, we can see that along the x dimension,
the potential is a double-well, whereas along the y and z dimensions,
we've just got a harmonic potential. So, we should expect that x is the slow
degree of freedom, whereas the system should equilibrate rapidly along y and z.
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
xx, yy = np.meshgrid(np.linspace(-2,2), np.linspace(-3,3))
zz = 0 # We can only visualize so many dimensions
ww = 5 * (xx-1)**2 * (xx+1)**2 + yy**2 + zz**2
c = plt.contourf(xx, yy, ww, np.linspace(-1, 15, 20), cmap='viridis_r')
plt.contour(xx, yy, ww, np.linspace(-1, 15, 20), cmap='Greys')
plt.xlabel('$x$', fontsize=18)
plt.ylabel('$y$', fontsize=18)
plt.colorbar(c, label='$E(x, y, z=0)$')
plt.tight_layout()
import simtk.openmm as mm
def propagate(n_steps=10000):
system = mm.System()
system.addParticle(1)
force = mm.CustomExternalForce('5*(x-1)^2*(x+1)^2 + y^2 + z^2')
force.addParticle(0, [])
system.addForce(force)
integrator = mm.LangevinIntegrator(500, 1, 0.02)
context = mm.Context(system, integrator)
context.setPositions([[0, 0, 0]])
context.setVelocitiesToTemperature(500)
x = np.zeros((n_steps, 3))
for i in range(n_steps):
x[i] = (context.getState(getPositions=True)
.getPositions(asNumpy=True)
._value)
integrator.step(1)
return x
Run Dynamics¶
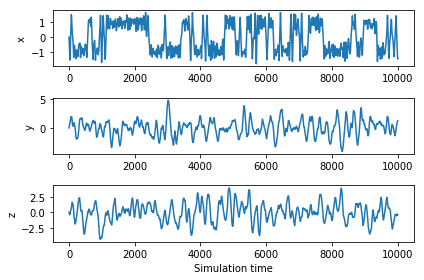
Okay, let's run the dynamics. The first plot below shows the x, y and z coordinate vs. time for the trajectory, and
the second plot shows each of the 1D and 2D marginal distributions.
trajectory = propagate(10000)
ylabels = ['x', 'y', 'z']
for i in range(3):
plt.subplot(3, 1, i+1)
plt.plot(trajectory[:, i])
plt.ylabel(ylabels[i])
plt.xlabel('Simulation time')
plt.tight_layout()
Note that the variance of x is much lower than the variance in y or z, despite its bi-modal distribution.
Fit tICA and PCA models¶
from msmbuilder.decomposition import tICA, PCA
tica = tICA(n_components=1, lag_time=100)
pca = PCA(n_components=1)
tica.fit([trajectory])
pca.fit([trajectory])
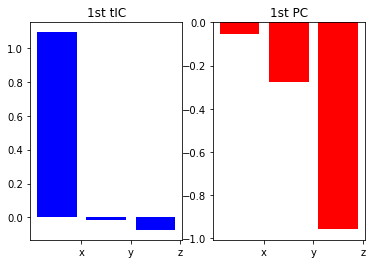
See what they find¶
plt.subplot(1,2,1)
plt.title('1st tIC')
plt.bar([1,2,3], tica.components_[0], color='b')
plt.xticks([1.5,2.5,3.5], ['x', 'y', 'z'])
plt.subplot(1,2,2)
plt.title('1st PC')
plt.bar([1,2,3], pca.components_[0], color='r')
plt.xticks([1.5,2.5,3.5], ['x', 'y', 'z'])
plt.show()
print('1st tIC', tica.components_ / np.linalg.norm(tica.components_))
print('1st PC ', pca.components_ / np.linalg.norm(pca.components_))
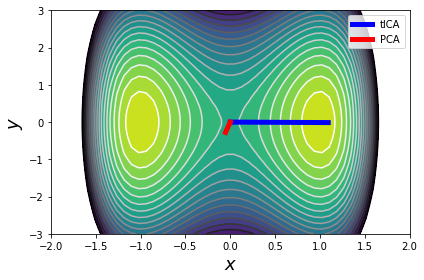
Note that the first tIC "finds" a projection that just resolves the x coordinate, whereas PCA doesn't.
c = plt.contourf(xx, yy, ww, np.linspace(-1, 15, 20), cmap='viridis_r')
plt.contour(xx, yy, ww, np.linspace(-1, 15, 20), cmap='Greys')
plt.plot([0, tica.components_[0, 0]],
[0, tica.components_[0, 1]],
lw=5, color='b', label='tICA')
plt.plot([0, pca.components_[0, 0]],
[0, pca.components_[0, 1]],
lw=5, color='r', label='PCA')
plt.xlabel('$x$', fontsize=18)
plt.ylabel('$y$', fontsize=18)
plt.legend(loc='best')
plt.tight_layout()