Python source code: [download source: plot_chord.py]
from msmbuilder.example_datasets import FsPeptide
import numpy as np
import mdtraj as md
import msmexplorer as msme
from msmexplorer.utils import make_colormap
# # Load Fs Peptide Data
trajs = FsPeptide().get().trajectories
# Compute Hydrogen Bonding Residue Pairs
baker_hubbard = md.baker_hubbard(trajs[0])
top = trajs[0].topology
pairs = [(top.atom(di).residue.index, top.atom(ai).residue.index)
for di, _, ai in baker_hubbard]
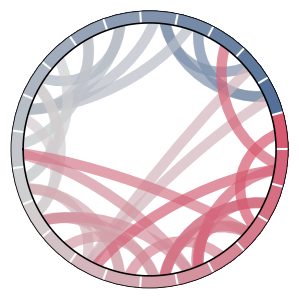
# Create Hydrogen Bonding Network
hbonds = np.zeros((top.n_residues, top.n_residues))
hbonds[list(zip(*pairs))] = 1.
# Make a Colormap
cmap = make_colormap(['rawdenim', 'lightgray', 'pomegranate'])
# Plot Chord Diagram
msme.plot_chord(hbonds, cmap=cmap)