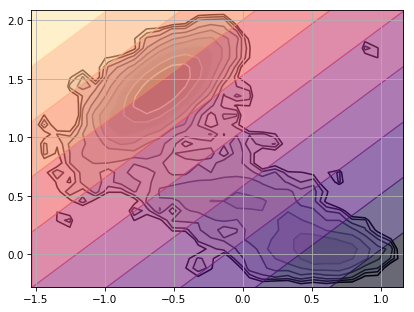
Python source code: [download source: plot_decomp_grid.py]
from msmbuilder.example_datasets import MullerPotential
from msmbuilder.decomposition import tICA
from msmbuilder.cluster import MiniBatchKMeans
from msmbuilder.msm import MarkovStateModel
import numpy as np
import msmexplorer as msme
rs = np.random.RandomState(42)
# Load Fs Peptide Data
trajs = MullerPotential().get().trajectories
# Perform Dimensionality Reduction
tica_model = tICA(lag_time=2, n_components=2)
tica_trajs = tica_model.fit_transform(trajs)
# Perform Clustering
clusterer = MiniBatchKMeans(n_clusters=12, random_state=rs)
clustered_trajs = clusterer.fit_transform(tica_trajs)
# Construct MSM
msm = MarkovStateModel(lag_time=2)
assignments = msm.fit_transform(clustered_trajs)
# Plot Free Energy
data = np.concatenate(trajs, axis=0)
pi_0 = msm.populations_[np.concatenate(assignments, axis=0)]
ax = msme.plot_free_energy(data, obs=(0, 1), n_samples=100000, pi=pi_0,
random_state=rs)
# Plot tICA Projection
msme.plot_decomp_grid(tica_model, ax=ax, alpha=0.6)