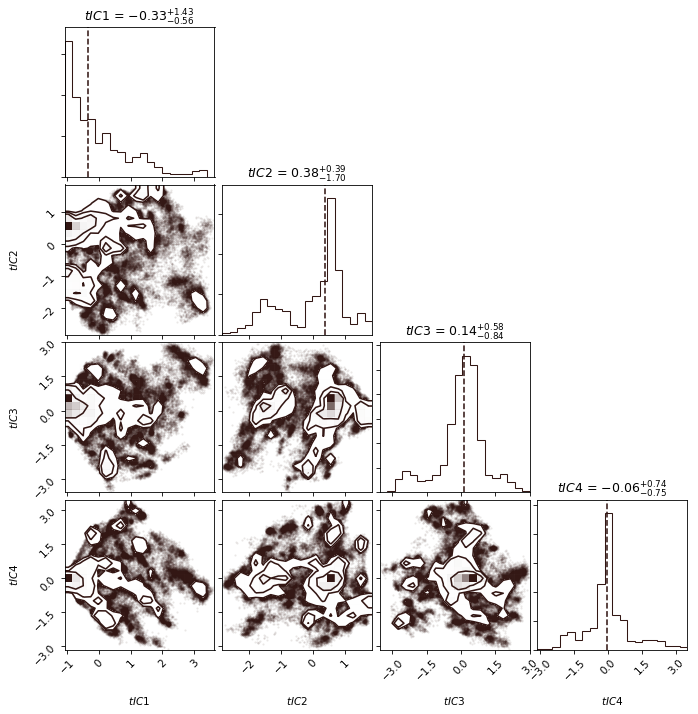
Python source code: [download source: plot_histogram.py]
from msmbuilder.example_datasets import FsPeptide
from msmbuilder.featurizer import DihedralFeaturizer
from msmbuilder.decomposition import tICA
import numpy as np
import msmexplorer as msme
# Load Fs Peptide Data
trajs = FsPeptide().get().trajectories
# Extract Backbone Dihedrals
featurizer = DihedralFeaturizer(types=['phi', 'psi'])
diheds = featurizer.fit_transform(trajs)
# Perform Dimensionality Reduction
tica_model = tICA(lag_time=2, n_components=4)
tica_trajs = tica_model.fit_transform(diheds)
# Plot Histogram
data = np.concatenate(tica_trajs, axis=0)
msme.plot_histogram(data, color='oxblood', quantiles=(0.5,),
labels=['$tIC1$', '$tIC2$', '$tIC3$', '$tIC4$'],
show_titles=True)